DeepCure is harnessing the power of AI-driven drug discovery to accelerate the development of novel, first-in-class precision therapeutics in areas of unmet medical need.
Our experienced scientists and engineers optimize all phases of drug-discovery, including early hit identification, hit-to-lead, lead optimization, patent strategy, and preparation for IND filing, yielding shorter timelines to develop the highest-quality drug candidates.
We are using AI-driven drug discovery to ensure better molecules and faster cures. Our mission is to discover, develop and deliver to patients novel, small molecule drugs that were previously undruggable or undiscovered until our technology came along.
Binding Modeling
Hypothesis Generation

One of the most critical components of our drug discovery pipeline, and one that differentiates us from the rest of the AI drug discovery companies, is our ability to create causal, data-driven, human-interpretable hypotheses for a wide range of targets.
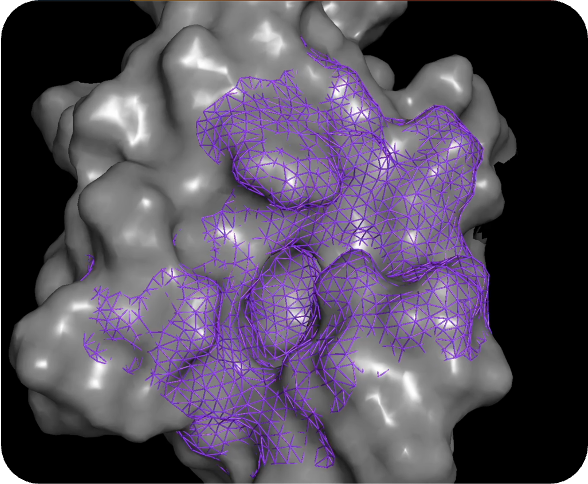
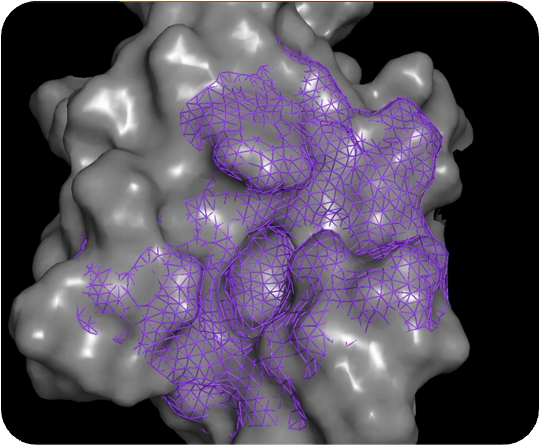
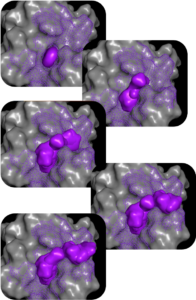
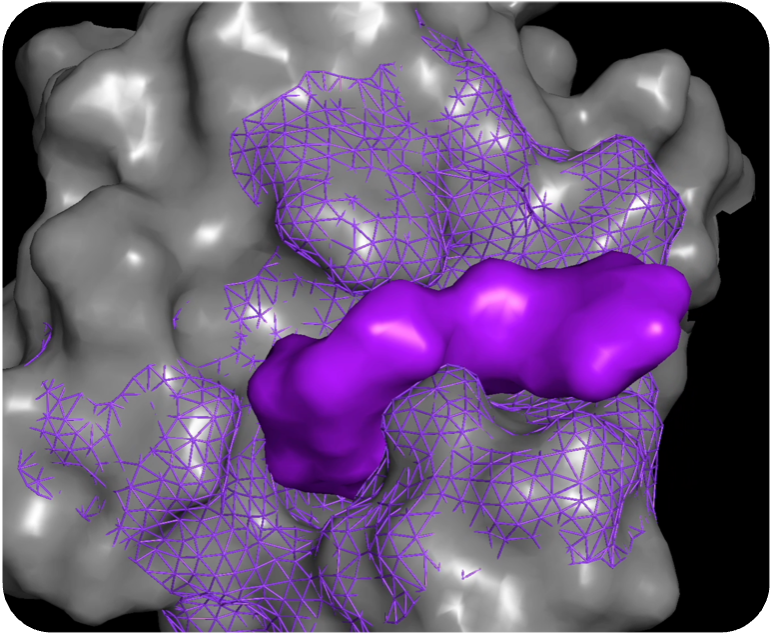
PocketExpander™
Discovering novel binding modes is the first step towards finding novel therapies
For most therapeutic targets of interest today, there is either no data, limited data, or biased data. For that reason, we start by generating a hypothesis from scratch, using only information about the approximate binding region that gives rise to the desired phenotype. Using PocketExpander™, our proprietary pocket mapping tool that leverages AI/ML as well as comp Chem methods, we are able to identify potential relevant binding modes. The output is then used as an initial binding hypothesis that is fed into MolGen™. The data obtained from synthesis and physical testing of molecules predicted by MolGen™ are then used to refine our hypothesis using causal analysis.
Causal Analysis
Machine Learning methods usually used for drug discovery are inherently focused on correlations. Since data sets around binding are often very small and highly biased, these methods will often get trapped by the bias and be unable to discover truly novel chemical matter. Our Causal ML methods for Hypothesis Generation analyzes the 3-dimensional structure from a physics-based perspective and uses causal analysis to find just the interactions relevant for binding in an unbiased way; thereby, unlocking new indications and safety profiles.
Medicinal chemists engage in a conversation with explainable models
On top of the strength of the causal hypothesis, it is designed to be human-interpretable, allowing scientists to make updates or experiment further in novel binding/selectivity modes. Causal features can then be shown as heatmaps in 3D (or mapped to 2D) and can be reviewed by medicinal and computational chemists to identify irregularities or gaps and customize them to explore additional interesting regions or interactions. This allows for a feedback cycle that ensures we don’t blindly follow the ML algorithm or a chemist’s intuition.

TCP-Aware Drug Design
MolGen™
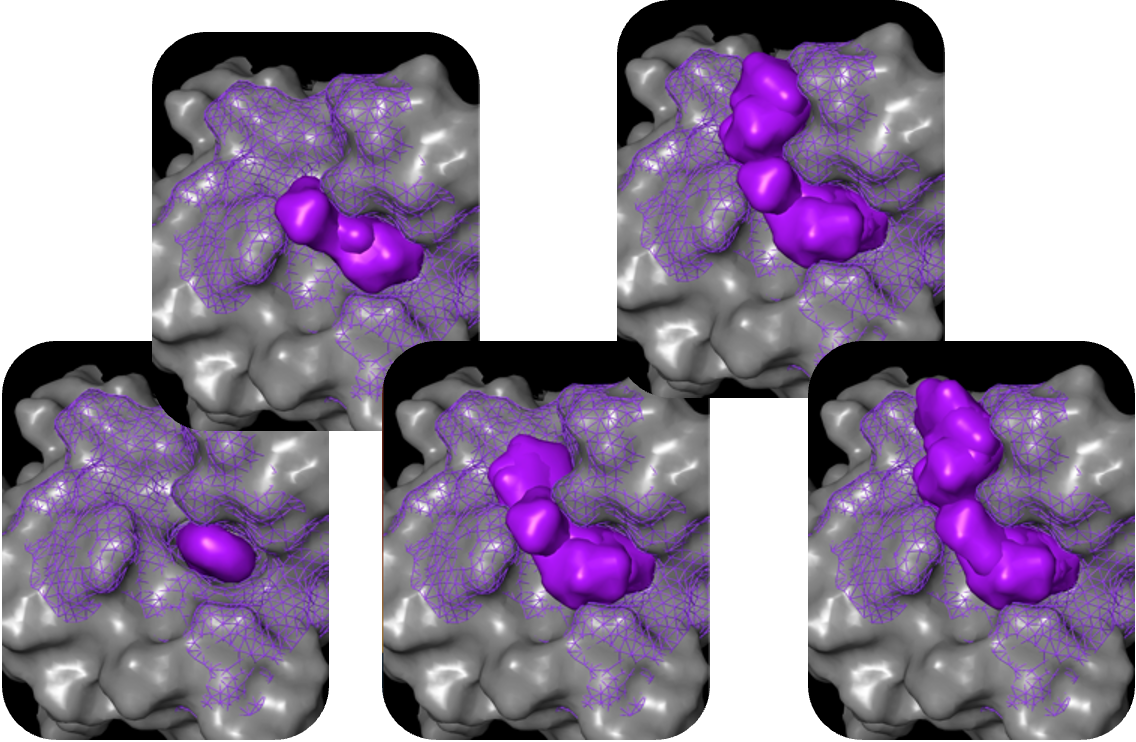
Our patent-pending molecular generation tool, MolGen™, is used to design novel, diverse compounds that are readily synthesizable and fit the desired TCP for a given drug program.
The molecular generation process leverages state of the art deep reinforcement learning (RL) to construct synthesizable molecules that efficiently capture all causal interactions for binding and selectivity while maintaining the desired ADME-tox profile.
MolGen™ utilizes hundreds of workers, choreographed in a unified pipeline (leveraging distributed training, hyperparameter optimization, GPU instances, and more), to improve the output quality of molecules as quickly as possible.
Prospective Evaluation of Accuracy
Multiparameter Optimization (MPO)
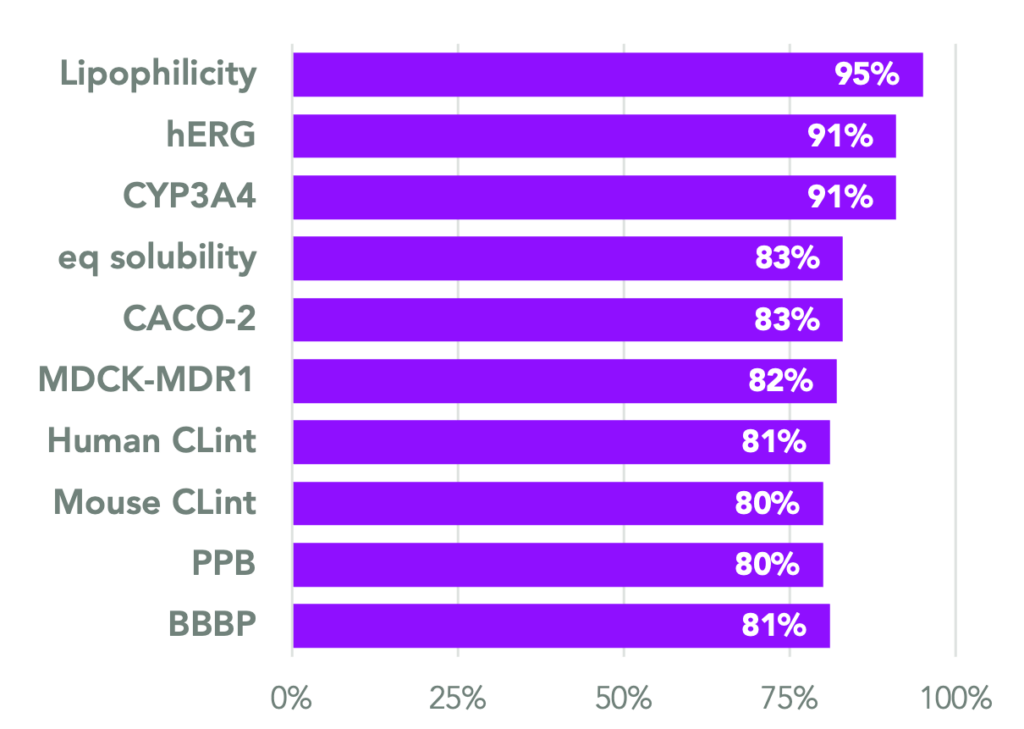
In parallel to our binding and selectivity hypothesis generation models, we have trained several state-of-the-art models (DeepPropR™), leveraging both public and proprietary data sources, to predict important pharmacological properties (ADME-Tox). Our DeepPropR™ models are frequently validated experimentally on molecules we synthesize to ensure an accurate measurement of the models’ performance and generalizability.
The figure (left) depicts the accuracy of our ten most used DeepPropR™ models from a prospective evaluation. Most of these accuracy data include results from at least 200 compounds that were not part of the training set but were generated from our internal pipeline and subsequently evaluated via physical testing. DeepPropR™models are also available for on-the-fly inference at scale using our custom-built Molecular Property Service (learn more in our relevant blog post, here).
Robotic Synthesis & Automated Physical Testing
Molecular Foundry
A large number of potentially outstanding compounds designed for a given drug discovery program are tossed as a result of decisions that led to them either not being synthesized or only partially characterized due to long timelines and high costs of manual synthesis and assays.
DeepCure’s Automated Molecular Foundry seeks to reduce design-build-test-learn cycle times and cost without limiting novelty and diversity of compounds via a fully integrated platform that includes robotic automation software, robotic synthesis, and assay automation.